Mapping how genes causally shape biology.
By testing every gene across all human tissues using large-scale Mendelian randomization and colocalization across thousands of diseases and biomarkers, we generate billions of causal data points linking genes to biological outcomes.
Our causally annotated data — and the AI models trained on it — create a holistic map that deepens our understanding of human biology, accelerating the discovery of novel drug targets and improving clinical trial design across therapeutic areas.
Causality in drug development
The challenge in drug development is that clinical efficacy cannot be predicted. Success rates, particularly in phase 2 when efficacy is first tested, are below 30%.
The gold standard for establishing causality is the prospective, randomized, placebo-controlled trial, which is necessary for drug testing. Outside of biomedicine, due to time and cost constraints, causality can only be investigated prospectively to a limited extent.
Thus, statistical methods have been developed to derive causal relationships from observational data retrospectively, known as causal inference.
.png)
Human genetics: natural clinical trial
The impact of the Human Genome Project on drug research is becoming increasingly evident. Retrospective analyses have shown that compounds binding to drug targets whose genes are associated with the disease in genome-wide association studies (GWAS) are significantly more successful in phase 2.
This genetic support is already being intensively utilized. In 2021, two-thirds of the drugs approved by the United States Food and Drug Administration (FDA) had genetic support. The analysis of GWAS data with causal AI to verify drug targets and predict clinical efficacy is simply a logical progression.
.png)
Mendelian randomization
Mendelian randomization (MR) is a causal inference method that leverages instrumental variables (IV), specifically genetic variants, as proxies for exposures of interest.
Because these genetic variants are randomly allocated at conception according to Mendel’s laws, they are generally independent of confounding factors that can bias observational studies—assuming the variant influences the outcome only through the exposure. This allows for estimation of the causal effect between an exposure and an outcome.
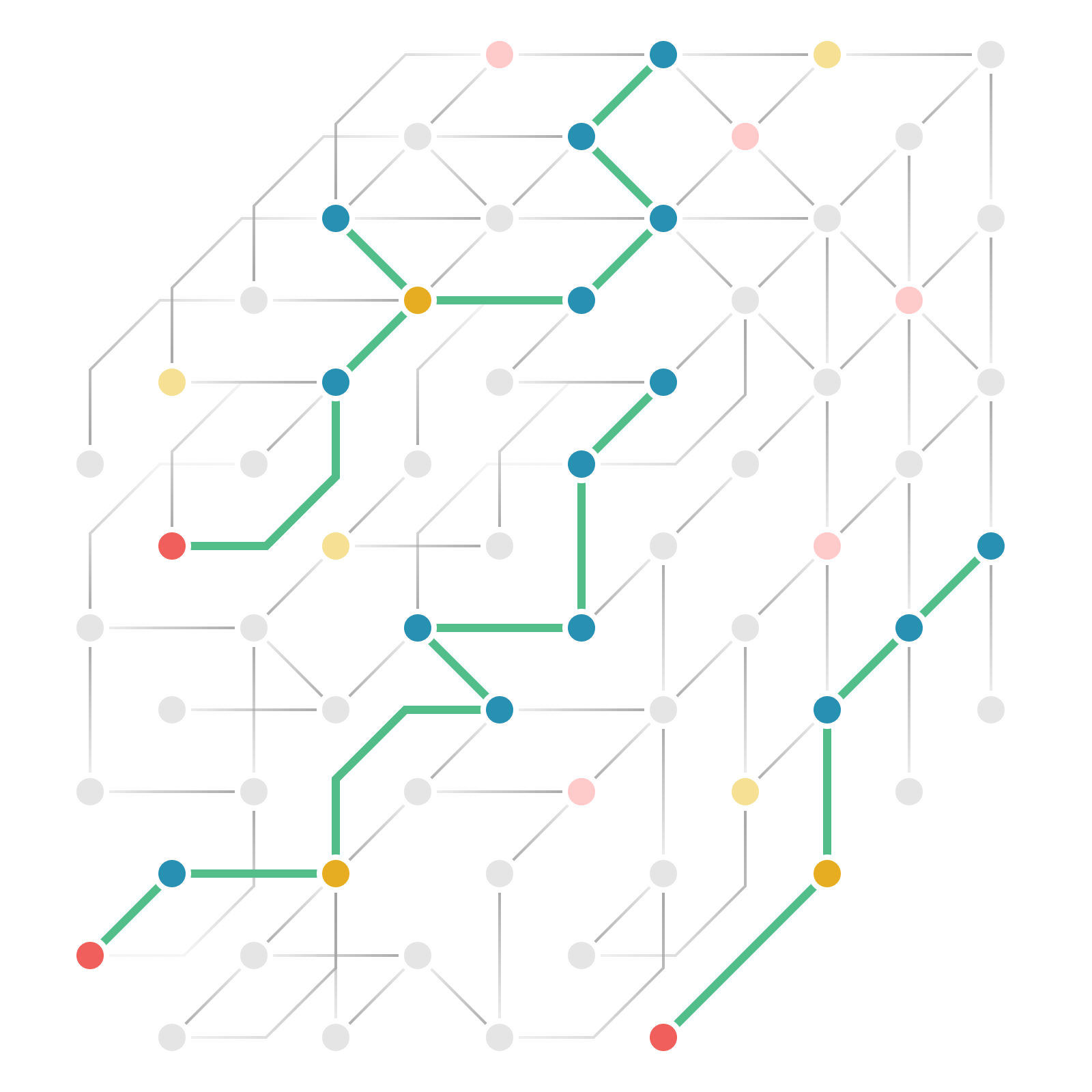
Mapping how genes causally shape biology
To systematically apply MR at scale, biotx.ai curated a comprehensive catalog of thousands of genome-wide association studies, encompassing more than 25 million cases across more than 3,000 diseases. Using this dataset, the company conducted MR analyses on all 23k human genes and over one million omics markers across 60 tissues for each disease.
Our causally annotated data — and the AI models trained on it — create a holistic map that deepens our understanding of human biology, accelerating the discovery of novel drug targets and improving clinical trial design across therapeutic areas.
This genetic support is already being intensively utilized. In 2021, two-thirds of the drugs approved by the United States Food and Drug Administration (FDA) had genetic support. The analysis of GWAS data with causal AI to verify drug targets and predict clinical efficacy is simply a logical progression.